Research in the Department of Biochemistry and Molecular Biology falls into three general areas: Gene Expression, Cellular Biochemistry and Structural Biology. The three groups are highly interactive and collaborative, both within and between the focus areas.
STRUCTURAL BIOLOGY
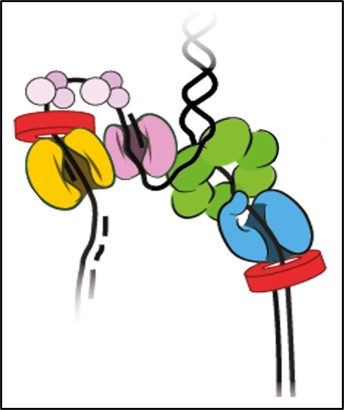
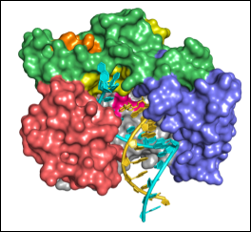
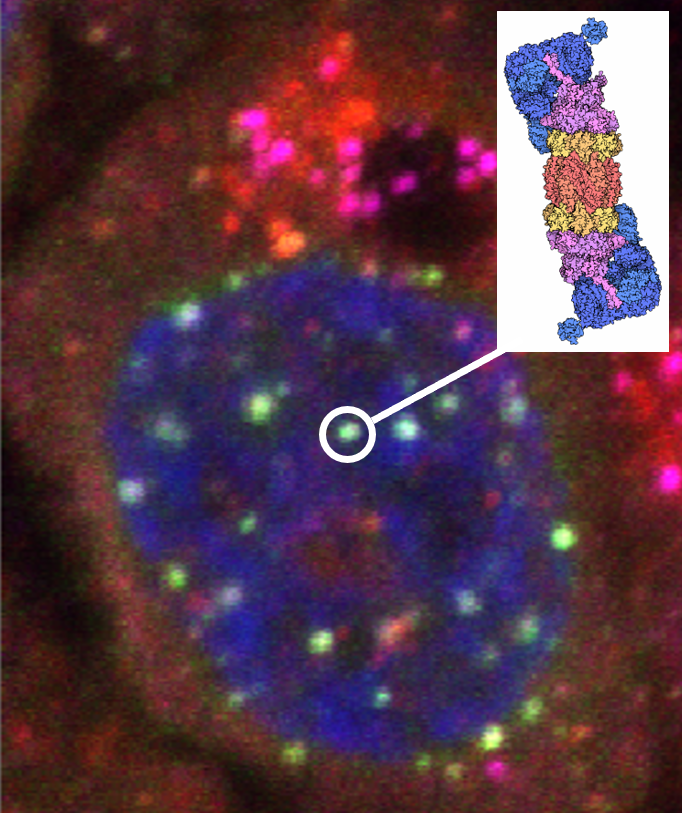
If a picture is worth a thousand words, then “seeing” the structure of a biomolecule or its complex is invaluable in understanding biological function. Structural biology research in the department seeks to unravel the molecular basis for how DNA is compacted in chromatin structure for packaging in a cell’s nucleus, how the genetic information in DNA is read by proteins during gene expression, how the genetic information encoded in RNAs are replicated in viruses, how the interactions among proteins are involved in the cell cycle, how instability in proteins lead to aggregation, and how proteins can be reengineered to be more stable or perform new functions.
Structural Biology
If a picture is worth a thousand words, then “seeing” the structure of a biomolecule or its complex is invaluable in understanding biological function. Structural biology research in the department seeks to unravel the molecular basis for how DNA is compacted in chromatin structure for packaging in a cell’s nucleus, how the genetic information in DNA is read by proteins during gene expression, how the genetic information encoded in RNAs are replicated in viruses, how the interactions among proteins are involved in the cell cycle, how instability in proteins lead to aggregation, and how proteins can be reengineered to be more stable or perform new functions.
GENE EXPRESSION
To study gene expression is to understand how genetic information orchestrates the molecular functions of life. Research topics on gene expression in the BMB program include DNA structure, chromatin organization and dynamics, transcription and translation regulatory control. Our work encompasses investigations that zoom into the atomic level or expand out to the organismal level, aided by state-of-the-art technologies. To understand gene expression is fundamental to know how a cell functions, how cells differentiate into complex organisms, and how misregulation can lead to human diseases.
Gene Expression
To study gene expression is to understand how genetic information orchestrates the molecular functions of life. Research topics on gene expression in the BMB program include DNA structure, chromatin organization and dynamics, transcription and translation regulatory control. Our work encompasses investigations that zoom into the atomic level or expand out to the organismal level, aided by state-of-the-art technologies. To understand gene expression is fundamental to know how a cell functions, how cells differentiate into complex organisms, and how misregulation can lead to human diseases.
CELLULAR BIOCHEMISTRY
Cellular biochemistry focuses on the signaling processes that permit the elaborate cell-cell communication required in a multicellular organism. Many research groups are currently studying cellular biochemistry, focusing their research on the rapid changes in cytoskeletal proteins, protein kinases, membrane lipids, and small ions that accompany extracellular signals. These groups utilize a broad array of biochemical techniques including fluorescence and electron microscopy, gene expression, and electrophysiology to characterize these fundamental biological processes.
Cellular Biochemistry
Cellular biochemistry focuses on the signaling processes that permit the elaborate cell-cell communication required in a multicellular organism. Many research groups are currently studying cellular biochemistry, focusing their research on the rapid changes in cytoskeletal proteins, protein kinases, membrane lipids, and small ions that accompany extracellular signals. These groups utilize a broad array of biochemical techniques including fluorescence and electron microscopy, gene expression, and electrophysiology to characterize these fundamental biological processes.
James Bamburg
- Apoptosis/Immortalization
- Cytoskeleton
- Signal Transduction
All mediators of major known dementias (amyloid-β in Alzheimer’s, α-synuclein in Parkinson’s Dementia and the HIV gp120 protein in AIDS Dementia) induce cofilin-actin rods via the same pathway. This pathway requires NOX and PrPC as well as cytokine/ Chemokine receptors CXCr4 or CCR5. We are determining if orally available peptides with the ability to antagonize CXCR4 & CCR5 receptors and block rod formation can protect synapses. Website: James Bamburg‘s directory listing.
Soham Chanda
- Electrophysiology
Neurons in the human brain communicate with each other via specialized structures called synapses, that transmit and process information in a neural network. Our lab studies various cellular parameters that regulate synaptic transmission in human neurons using synaptic imaging and electrophysiology. For more information, please visit: https://sites.google.com/view/sohamchanda
Chaoping Chen
- Retrovirus Assembly and Budding
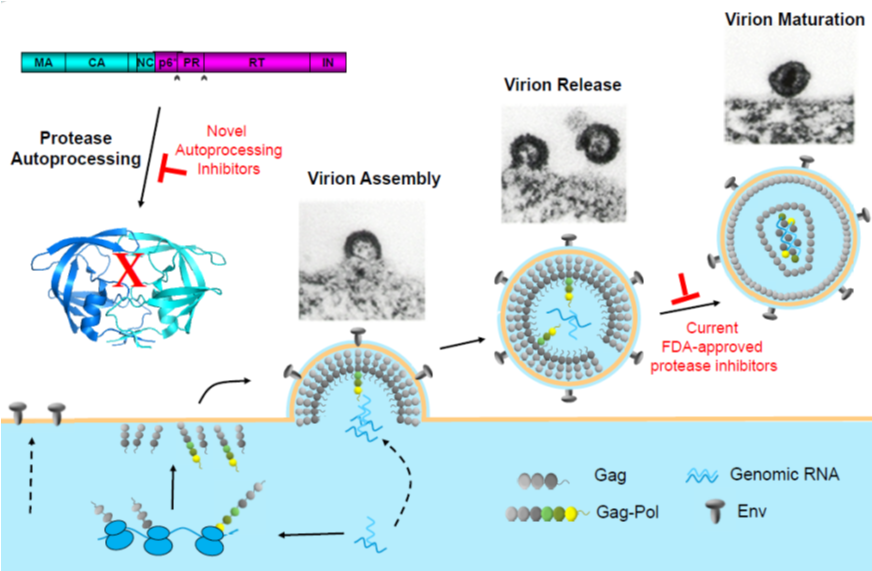
The HIV-1 protease is initially synthesized as part of a polyprotein precursor (Gag-Pol) whose autoprocessing is responsible for liberation of free mature protease in a highly regulated manner. We are interested in the underlying molecular and cellular mechanism and its contribution to drug resistance development. Our goal is to develop novel antivirals to help combat the ongoing AIDS epidemic. Website: Chaoping Chen‘s directory listing.
Robert Cohen
- Ubiquitin metabolism and ubiquitin-mediated signaling
Post-translational modifications by the protein ubiquitin and its diverse signaling functions are regulated by hundreds of proteins that mediate ubiquitin conjugation, recognition, and deconjugation. Our work aims to understand how different forms of ubiquitin signals are recognized, and to develop tools that can be used to probe ubiquitin dynamics in live cells. Website: http://yaocohenlab.bmb.colostate.edu
Jennifer DeLuca
- Mitosis
We study the fundamental biological process of mitotic cell division. The goals of our research are to understand the molecular mechanisms that ensure mitotic chromosome segregation fidelity and to determine how oncogenic signaling leads to chromosome mis-segregation. Website: https://delucalab.colostate.edu/
Steven Markus
- Cytoskeleton
The focus of the Markus lab is to understand how cellular cargos are delivered to the right place at the right time. Intracellular transport is a critical feature of cell physiology, and is largely mediated by a small group of molecular machines called motor proteins. In addition to determining how these motors are regulated to achieve appropriate transport, we are also focused on understanding how mutations in them lead to a variety of devastating human diseases, including developmental brain diseases, and motor neuron diseases. Website: https://markuslab.colostate.edu/file/Home.html
Erin Osborne Nishimura
Embryos develop through the regulated expression of many genes. Our lab studies how global gene expression impacts the development of embryonic as well as adult cells. Genes influence how cells transform into different cell types such as neurons or intestinal cells and regulate the distinct characteristics each cell will adopt. This work leverages high throughput sequencing, high-resolution imaging, and computational analyses within animal model systems. This work yields insight into the fields of cancer biology and human developmental diseases. Website: https://onishlab.colostate.edu/
Santiago Di Pietro
Our laboratory studies how cells take up materials via endocytosis and the biogenesis of specialized intracellular compartments in skin cells and platelets. Our research elucidates both normal physiology and developmental processes as well as disease mechanisms. Projects apply a range of experimental approaches from CRISPR/Cas9 gene editing, mouse and yeast genetics, and various microscopy modalities, to protein biochemistry and biophysics. For more information, please visit: Santiago Di Pietro‘s directory listing.
Tom Santangelo
We employ archaeal systems to investigate the regulation of fundamental processes (physiology, transcription, DNA replication, recombination) using a combination of biochemistry, NGS, genetic and microbiology techniques. Our diverse research portfolio explores the processes that permit life in the extremes and details shared regulatory strategies employed throughout the Archaea. For more information, please visit our website: http://santangelolab.colostate.edu/
Tim Stasevich
The Stasevich lab develops state-of-the-art technology to image and quantify gene regulatory dynamics with single molecule resolution in living cells. For more information, please visit: http://sites.bmb.colostate.edu/stasevichlab/
Tingting Yao
Maintaining the balance of protein synthesis and degradation, proteostasis, is central to a healthy cell. As cells age, their ability to maintain a healthy proteome declines, which contributes to a variety of age-related diseases. The ubiquitin-proteasome pathway and autophagy are the two degradation pathways that play critical roles in proteostasis. We aim to understand the mechanics of proteasome degradation and its role in regulating proteostasis in response to stress. Website: https://wp.natsci.colostate.edu/yaolab/
John Purdy
- Category A
- Category B
This is the template card for an individual. https://www.natsci.colostate.edu
John Purdy
- Category A
- Category B
This is the template card for an individual. https://www.natsci.colostate.edu
John Purdy
- Category A
- Category B
This is the template card for an individual. https://www.natsci.colostate.edu
Robert Cohen
- Analysis of Macromolecular Assemblies
Post-translational modifications by the protein ubiquitin and its diverse signaling functions are regulated by hundreds of proteins that mediate ubiquitin conjugation, recognition, and deconjugation. Our work aims to understand how different forms of ubiquitin signals are recognized, and to develop tools that can be used to probe ubiquitin dynamics in live cells. Website: http://yaocohenlab.bmb.colostate.edu
Jeffrey Hansen
- Analysis of Macromolecular Assemblies
- Protein Folding
- Analytical Ultracentrifugation
Chromatin—the genetic material of eukaryotes—consists of half DNA and half histone proteins. Our laboratory uses biochemical and biophysical methods to investigate the structure of chromatin and how chromatin structure relates to chromatin function. For more information, please visit: Jeffrey Hansen‘s directory listing.
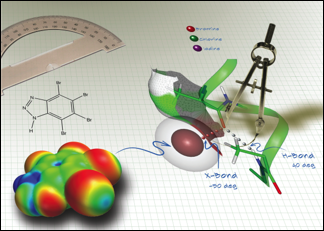
P. Shing Ho
- X-Ray Crystallography
Life has evolved using a very limited number of chemical elements. Our laboratory exploits the halogen bond (XB) to design new protein-protein interactions, stabilize intrinsically disordered proteins, and engineer new enzyme catalysts. We also develop computational tools to use XBs in drug design.
Website: P. Shing Ho‘s directory listing.
Olve Peersen
- Analysis of Macromolecular Assemblies
- X-Ray Crystallography
Our research is focused on the mechanisms of positive strand RNA virus replication, with a focus on their RNA-dependent RNA polymerases and the molecular mechanisms underlying the classically low fidelity of these enzymes that give rise to high virus mutation rates. Our experimental approaches are focused on protein biochemistry, structural biology, and rapid stopped-flow and quench-flow kinetics. Website: Olve Peersen‘s directory listing.
Eric Ross
- Amyloid Fibril Formation
Protein aggregation is associated with a wide variety of human diseases, including Alzheimer’s Disease and Amyotrophic Lateral Sclerosis (ALS). Our lab uses yeast prions as a model to study the causes and consequences of protein aggregation. For more information, please visit: Eric Ross‘s directory listing.
Grant Schauer
- Analysis of Macromolecular Assemblies
DNA replication is carried out by the replisome, a multiprotein machine. Our laboratory studies how the replisome and associated factors coordinate high-fidelity replication in the face of obstacles. For more information, please visit: Grant Schauer‘s directory listing.
Laurie Stargell
- Reconstitution of Chromatin; Chromatin Structure and the Regulation of Gene Expression
- Mechanisms of Transcriptional Initiation by RNA Polymerase II
Laurie Stargell‘s directory listing.
Tim Stasevich
- Regulation of transcription and translation at the single cell level
The Stasevich lab develops state-of-the-art technology to image and quantify gene regulatory dynamics with single molecule resolution in living cells. For more information, please visit: http://sites.bmb.colostate.edu/stasevichlab/
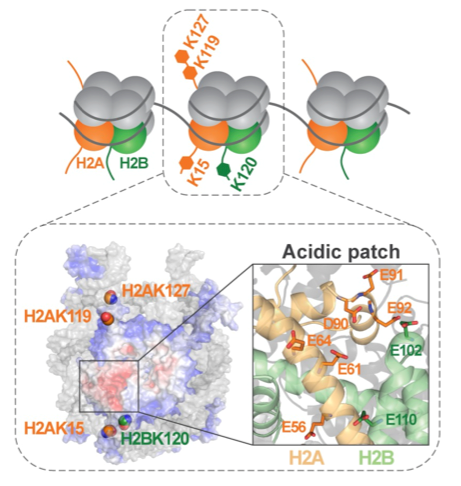
Tingting Yao & Robert Cohen
- Regulation of chromatin structure and dynamics by histone ubiquitination
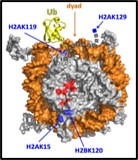
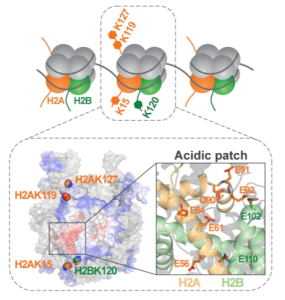
Post-translational modifications of histone proteins in chromatin play a major role in epigenetic mechanisms. We study ubiquitination of histones, which depends on the type of histone as well as the specific lysine to which ubiquitin is attached; histone ubiquitination signals a variety of consequences that include transcription activation, transcription repression, and DNA damage repair. Website: https://wp.natsci.colostate.edu/yaolab/
Erin Osborne Nishimura
- Regulation of transcription and translation at the single cell level
Embryos develop through the regulated expression of many genes. Our lab studies how global gene expression impacts the development of embryonic as well as adult cells. Genes influence how cells transform into different cell types such as neurons or intestinal cells and regulate the distinct characteristics each cell will adopt. This work leverages high throughput sequencing, high-resolution imaging, and computational analyses within animal model systems. This work yields insight into the fields of cancer biology and human developmental diseases. Website: https://onishlab.colostate.edu/
Tom Santangelo
We employ archaeal systems to investigate the regulation of fundamental processes (physiology, transcription, DNA replication, recombination) using a combination of biochemistry, NGS, genetic and microbiology techniques. Our diverse research portfolio explores the processes that permit life in the extremes and details shared regulatory strategies employed throughout the Archaea. For more information, please visit our website: http://santangelolab.colostate.edu/
Grant Schauer & Jeffrey Hansen
- Reconstitution of Chromatin; Chromatin Structure and the Regulation of Gene Expression
Chromatin contains epigenetic histone modifications that are critical to defining the transcription profile of a cell. Our laboratories study how chromatin is regulated during DNA replication, and how chromatin structure regulates transcription machinery. For more information, please visit: Grant Schauer‘s and Jeffrey Hansen‘s directory listings.
Soham Chanda
Neurons in the human brain communicate with each other via specialized structures called synapses, that transmit and process information in a neural network. Our lab studies various cellular parameters that regulate synaptic transmission in human neurons using synaptic imaging and electrophysiology. For more information, please visit: https://sites.google.com/view/sohamchanda
Jeffrey Hansen
- Reconstitution of Chromatin; Chromatin Structure and the Regulation of Gene Expression
Chromatin—the genetic material of eukaryotes—consists of half DNA and half histone proteins. Our laboratory uses biochemical and biophysical methods to investigate the structure of chromatin and how chromatin structure relates to chromatin function. For more information, please visit: Jeffrey Hansen‘s directory listing.
P. Shing Ho
- X-Ray Crystallography
A cell’s genetic information is coded not only in the bases of DNA, but also by chemical modifications to these bases (epigenetic modifications). Our laboratory studies how 5-hydroxylmethylcytosine is recognized by proteins that help distinguish vertebrate from invertebrate organisms.
Website: P. Shing Ho‘s directory listing