Mentors and Projects Available to Choose From Summer 2025
Miriam Braunstein
Department of Microbiology,Immunology and Pathology
Research Profile
Research Interests:
My laboratory studies the basic biology and pathogenesis of mycobacterial pathogens, including Mycobacterium tuberculosis and nontuberculous mycobacteria (NTM). We study the mycobacterial protein secretion pathways and the role they play in enabling intracellular survival of mycobacteria in macrophages. We also investigate novel therapeutics, including bacteriophage therapy, as treatments for drug resistant mycobacterial disease.
Mentoring Plan:
I am dedicated to mentoring trainees at all levels. At the start of the summer, I will meet with the student, and we will develop a list of agreed upon expectations for the summer research experience. I will then meet with the undergraduate student weekly to discuss their research project and progress. The student will also be assigned a ‘bench mentor’ who will meet daily with and advise the student in the lab. The student will participate in our weekly lab meetings and journal clubs. At the end of the summer, we will work with the student in writing an abstract on their summer research project and presenting their summer research at lab meeting.

Debbie Crans
University Distinguished Professor
Department of Chemistry and Cell and Molecular Biology Program
Research Profile
Research Interests:
As a biological chemist my laboratory develops new drugs for treatment of platinum-resistant glioblastoma (T98g) cell lines using intratumoral administration techniques. We have developed new vanadium-containing compounds that are effective anticancer agents, and we propose to study new potential compounds with similar activities. We have explored different administration techniques including addition of serum albumin that significantly improve the efficacy of the compounds in cell culture studies by modifying the mechanism of action.
Mentoring Plan:
To design new compounds as potential anticancer compounds, we prepare bulky hydrophobic ligands to coordinate to the vanadium atom. The objective is to design compounds for intertumoral administration which requires that the compounds are reactive but also survive administration and only react when injected into the tumor. We plan to design compounds with varying hydrolytic stability under the assay conditions and do so by the addition of steric bulk and increased hydrophobicity of the ligand bound to the metal ion. In addition to testing the anticancer effects of these compounds we propose testing their effects of respiration in mitochondria. The REU student will participate in weekly group meetings with both the chemical group and that of our biological collaborators (the Chicco group at CSU and the Lay group at the University of Sydney, Australia).
Recent reference of related work: “Vanadium(V) pyridine-containing Schiff base catecholate complexes are novel lipophilic, redox-active and selectively cytotoxic in glioblastoma (T98g) cells” Kateryna Kostenkova, Aviva Levina, Drew A. Walters, Heide A. Murakami, Peter A. Lay, Debbie C. Crans Chem. A Eur. Journal 2023 e202302271 (1 of 16) https://doi.org/10.1002/chem.202302271 Inside Cover was published. (Drew A. Walters is an undergraduate co-author)

Chaoping Chen
Department of Biochemistry & Molecular Biology
Research Profile
Research interests:
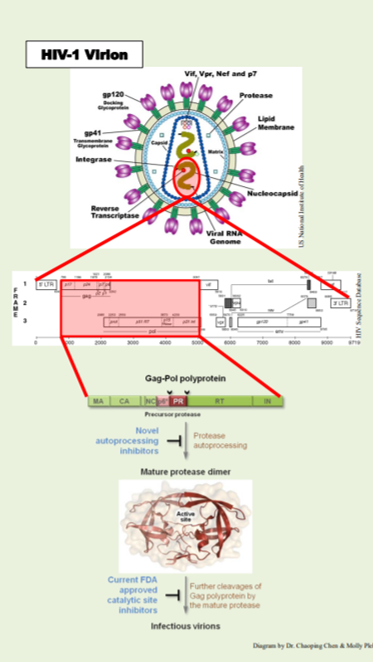
Retroviruses are enveloped RNA viruses, which comprise a large and diverse family including human immunodeficiency virus 1 (HIV-1), the pathogen that leads to AIDS. The laboratory focuses on characterization of the molecular and cellular mechanisms underlying the late stage of HIV-1 replication using a combination of biological imaging, cellular and molecular virology approaches.
Mentoring plan:
The REU fellow will learn basic molecular biology techniques including SDS-PAGE, western blotting and subsequent quantification analysis, as well as fluorescence imaging and quantification protocols specifically established for the phenotype assay, under the guidance of lab personnel. Results obtained will shed new lights into drug resistance mechanism. Through these research activities, the REU fellow will also develop skills of critical thinking, spoken and written communication, and being a productive team player.
Diego Krapf
Department of Electrical and Computer Engineering
Research Profile
Research Description:
Our lab focuses on the architecture of mammalian cells. The main expertise of the team is in single-molecule biophysics experiments at the nanometer scale. We place particular emphasis on the actin-based cytoskeleton and its role in the dynamic organization of mammalian cells. The nature of this field is highly interdisciplinary, crossing the boundaries of Engineering, Physics, and Biology. The techniques we use in our research consist of stochastic processes, single-molecule detection and tracking, and superresolution imaging.
Mentoring Plan:
Day-to-day mentoring: A postdoc will be assigned to the day-to-day mentoring of the REU participant.
Interactions with the PI and other group members: The PI will meet at least twice per week with the mentor and the REU participant to discuss project progress and plan the work, once in a project-specific meeting and once during weekly group meetings. The group meetings will provide an opportunity for the participant to discuss the progress with the whole group. In addition, the REU participant will have the opportunity to participate in a weekly group lunch and a weekly journal club. Our group organizes outings (i.e., to the Poudre River or a hike in the mountains) and we always organize one of these outings when we have lab visitors.
Mentor training: Before the start of the project, the PI will thoroughly discuss with the mentor the responsibilities of the REU participant, the training of the participant, expected time commitment, and expected outcomes. The PI will meet with the mentor weekly during the project to assess the participant’s progress, to rapidly address any challenges, and to set new goals if needed.
Expectations and goals: The REU participant is expected to work in the lab 40 hours per week. The student will be given journal articles to read, and these articles will be discussed with the mentor and the PI. The REU participant is also expected to become familiar with appropriate lab safety protocols (these are achieved via online training) and maintain an electronic notebook. Depending on the specific project assigned to the student, they will be expected to achieve one of the following sets of goals.
i)Be trained in imaging in a confocal microscope. Understand the limits of diffraction-limited imaging and how to perform super-resolution imaging. Collect sperm or spermatid samples from mouse. Fix cells and label them for imaging. Optimize labeling conditions for immunofluorescence. Along this project the participant will be guided to think critically of the results and to test specific hypotheses.
ii)Perform statistical analysis of trajectories obtained by single-particle tracking in living cells. Model different modes of motion. Perform numerical simulations of random walks as a model for the motion of molecules in living cells. In this project, the participant will receive training in statistical methods using MATLAB tools.
iii)Perform live cell imaging under physiological conditions. Culture cells for track force microscopy. Obtain calibrated track force microscopy measurements where the cell exerts forces under different environmental conditions. In this project the participant will employ statistical methods and test specific hypotheses.
Evaluation and feedback: During the last two weeks of the project, the PI will gather the participant’s feedback on his experience, discuss research interests beyond the 10-week program and talk about career options. The student will give a short oral presentation where they discuss their work.
Graham Peers
Department of Biology
Research Profile
Research interests:
My primary research interests are: photosynthetic efficiency, how photosynthesis is regulated in a dynamic environment and algal stress physiology. Researchers in the laboratory employ forward and reverse genetics, ‘omics approaches and detailed physiology to investigate these areas. We are particularly interested how these processes differ across the vast evolutionary diversity of algal and cyanobacterial clades.
Mentoring plan:
A senior level graduate student or postdoctoral scholar will participate in day-to-day mentoring with assistance from another graduate student and one research scientist. They will be responsible for teaching lab techniques and analyses. I will meet with the student(s) weekly both in lab meeting and one-on-one to discuss progress, identify roadblocks and also help in the lab when needed. Our goal will be to have the student(s) develop one figure (each) for a publication in a scientific journal.

Marcelo Melo
Department of Biochemistry and Molecular Biology
Research Profile
Research interests:
Our laboratory focuses on the use and development of computational tools to explore biochemical problems. The techniques we use range from software development to AI/ML models, and our interests connect the sub-atomic details of an enzymatic reaction to the regulatory mechanisms coordinating metabolic pathways. Blending mathematical and physical models with software development and data science, we explore the mechanisms behind molecular interactions and look for the atomic origins of biochemical transformations that decide a cell’s fate.
The hands on, day-to-day work involves performing computer simulations, gathering large scale datasets, combining biological, physical, and chemical information into our models, and creating AI/ML models to extract information and insight from terabytes of raw data.
We rely heavily on experimental data from collaborators and public repositories to create the foundations of simulations and tests new hypotheses. The research infrastructure ranges from local to national high performance computing clusters, and we will focus on using and publishing open source software packages and models.
Mentoring plan:
Students joining the lab for the REU program will be directly involved in hands-on research in computational biochemistry. Students can expect close supervision by myself and other lab members and will be expected to focus their work efforts on the project they join for the duration of the program.
Research Project: Mentoring will start by integrating the student to an ongoing research project. The broad range of techniques and problems explored in the lab provide a variety of opportunities for tailoring projects to student’s academic backgrounds and interests. This will allow us to optimize productivity (by leveraging the student’s prior experience) and learning opportunities (by aligning the lab’s and the student’s goals).
Academic and social integration: The student will join lab members when participating in departmental events (e.g., weekly seminars and social hours), and will join the lab’s weekly group meetings and project meetings. This will give the student multiple opportunities to learn about other research efforts, inside and outside the lab, and develop networking skills. When presenting their work, the student will also have chances to train presentation and discussion skills in an interdisciplinary environment.
Mentoring dynamics: The student can expect meetings with the PI at least twice per week, and direct access to the PI for training and supervision throughout the program. The student will have day-to-day interactions with lab members for support and discussions, and will be encouraged to explore their assigned project while developing independence and self reliance. Communication in a research lab is essential, and the student will be expected to discuss project issues with the PI and lab members, receive training and feedback, and reach out to the PI and lab members with questions and doubts throughout the program.
Work dynamics: The student will be expected to work solely on theory and computational tasks for the development of their project. This will involve reading and discussing scientific articles, implementing code or models in python/c++, training and deploying AI/ML models, preparing visualizations and reports, and presenting their work to the lab. The specifics will vary from project to project, and can be reshaped during the program to accommodate evolving needs. The project will balance a student’s past experience while allowing them to explore new tools and theories.
The student will receive specific training on relevant techniques (examples below) and will document the performed work to participate in scientific publications. While much of the work performed in the lab can be completed remotely through diverse computational resources, this training program – as well as true lab integration – can only occur with participation on site. Therefore, the student will be expected to spend at least 32h/week in the lab.
Computational Structural Biology: Projects in this area will involve learning how to prepare a molecular system for publicly available sequences and structures, how to perform molecular dynamics simulations, apply perturbations to the system to determine their response to physiological conditions, quantify physicochemical properties such as binding free energy, and estimate barriers to system dynamics. These techniques are used in academic basic science and industry applied research, in the broad fields of computational biology and computational chemistry, and help us understand the atomic origins of molecular interactions and behavior, and can ultimately explain the origins of a pathological condition or the mechanism of action of a new drug.
Data Science: Projects in this area will include exploring, combining, and preparing datasets for AI/ML model training, development of traditional and/or deep neural network models, integration of biochemical information and theoretical predictions into training loops, validation of models with experimental data, and visualization of high-dimensional datasets. These techniques are part of an evolving landscape of modern statistical analysis, and are being actively integrated in every aspect of basics science, industry research, and commercial activities. Here, they will help us extract information and detect patterns from large datasets of experiments and simulations, and can ultimately reveal connections between molecular properties and physiological states of cells, or properties of molecular systems that show relevant biochemical activity.

Tom Santangelo
Department of Biochemistry & Molecular Biology
Research Profile
Research interest:
We investigate the information processing systems and metabolism of microbes that thrive in extreme environments. Using a combination of genetic and biochemical techniques we aim to understand the evolution, mechanisms, and interplay of archaeal transcription, DNA repair, DNA replication, and epigenetic modifications. In short, we strive to understand how microbes can survive under conditions where most would quickly perish due to temperatures, pressure, pH or salinity extremes. We regularly employ >10 undergraduates and >7 graduate mentors that direct independent research efforts towards these aims.
Mentoring plan:
I am very familiar with the demands of the REU program, the mentoring philosophy and training format, and the need for assistance at the bench and with the totality of scientific training. I meet with REU students one-on-one each week; REU students present at weekly group meeting to solicit feedback and practice presentation skills. We also aim to socially incorporate REU students into our large group, an often overlooked yet critical feature of summer and scientific training. So much can be learned from informal gatherings and interactions that we aim to ensure full immersion in such.
Grant Schauer
Department of Biochemistry and Molecular Biology
Research Profile
Research Interests:
Challenges to DNA replication, called replication stress, pose major risks to genome fidelity. Our lab studies the molecular mechanisms of various replication-coupled machines that help achieve accurate and efficient chromosome duplication in the face of such stress. Current research topics include the replication checkpoint apparatus, replication-coupled translesion synthesis, and conflicts between replication forks with transcription bubbles and other R-loop proteins. We study these molecular mechanisms in detail with single-molecule fluorescence techniques, traditional biochemistry, genetics, and cell biology. For instance, we are studying The REU student will be given the choice between a few projects, depending on the current needs of our lab and on the interests of the student.
Mentoring plan:
My role as PI will be to meet with the student several times a week, tracking progress, helping with protocols and techniques, defining the research direction of the project as it evolves, and providing personalized input for growth as a scientist. Depending on the project chosen, students will be assigned to work directly with at least one graduate student, who will be their primary day-to-day mentor. Besides myself, our lab manager Alisa will assist incoming students in the laboratory. Alisa has over 20 years experience at the bench at CSU and has trained over 100 students in molecular biology, biochemistry, cell biology, and microscopy techniques.
My implicit expectations are that the student shows a sense of urgency and motivation, demonstrates extra effort towards learning new concepts and improving skills at the bench, and is willing to communicate in detail with myself and other lab members. The primary expected achievement is that the student learns to conduct experiments independently by the end of the summer, with the ultimate goal of a direct contribution on a peer-reviewed research article.

Chris Snow
Department of Chemical and Biological Engineering
Research Profile
Research interests:
The lab focuses on computer-guided protein and DNA engineering, particularly the engineering of crystals. Methods of particular interest include directed evolution, macromolecular crystallography, protein structure prediction, and new algorithms for reliable computational protein engineering. The lab works on designing and experimentally testing a new class of self-assembling crystals composed of both protein and DNA building blocks.
Mentoring plan:
The REU student will work within a cohesive subgroup in the lab that is dedicated to the design and experimental testing of new protein-DNA co-crystal assemblies. Thus, they will receive training from multiple mentors (3 experienced Ph.D. students). The student will also get to interact with a larger set of students in the group and participate in journal club, lab meetings, and optionally get more experience with protein production and purification by participating in the summer protein production assembly line. They will meet with the PI 1:1 regularly to supplement participation within the weekly subgroup and weekly group meeting.

Kathryn Wilsterman
Department of Biology
Research Profile
Research interests:
Current projects in the lab are focused on understanding how reproductive physiology deals with hypoxic environments at high elevations. We use deer mice from Nebraska and Mount Evans, Colorado (over 14,000 ft elevation) to understand both how hypoxia limits reproduction, and how physiology in high elevation animals has overcome these challenges through physiological adaptation. We are combining in vitro approaches and organismal physiology to ask how tissues like the ovaries and uterine lining respond to chronic hypoxia. Over the course of the summer, the student will gain experience working with rodent models, histological approaches to reproductive physiology, and organismal biology.
Mentoring plan:
The REU student will work within a team in the lab and receive training from multiple mentors, including two technicians in the lab. I will meet with the student weekly to lay out goals, revise summer-long outcomes, and identify opportunities for further development. The student will have the opportunity to interact with a larger set of students in the laboratory and on the floor, including several graduate students in my lab and others. The student will be encouraged to participate in summer journal clubs and lab meetings.

